Summary of the recent Nature Biotechnology article on “Transparency Tools in Gene Patenting for informing policy and practice.”
By Osmat A. Jefferson, Cambia, Australia & Queensland University of Technology, Brisbane, Australia
As the global intellectual property (IP) system grows and now impacts virtually all citizens, it is crucial that the means to understand these rights and their teachings, as well as their implications and scope become global public goods. To do so requires not only that the primary data is available freely and openly in a standardized and re-useable form, but that tools to visualize, analyse and model that data are similarly open and free public goods, adaptable to diverse needs and uses; this we call ‘transparency’.
Open web-based platforms that enable the aggregation, commentary and mapping of this knowledge by the community and that transcend any one jurisdiction or field of innovation, are also needed.
These imperatives have informed Cambia’s development of The Lens (an open access, autonomous web-based patent search facility), and in particular, its biological innovation capability, part of which is referred to here as the PatSeq facility (see box).
Gene patents are among the most contentious, opaque and poorly understood aspects of modern IP. Inventions and products or services that hinge on knowledge of or direct use of the sequences that make up genetic material, typically DNA, or the proteins that genes encode, are becoming common and very important. Societal concerns about the appropriateness of allowing patenting of these components of living systems have also become prominent.

Transparency in relation to the extent and scope of genetic inventions is critical, and has several important roles. First, transparency allows examiners to understand the invention and to determine if it meets patentability requirements. Second, it enables public access to, and use of, the invention concept to enact the patent ‘bargain’ or ‘compact’, thereby fostering follow-on inventions, cumulative innovation, and reducing duplication of effort. Third, transparency gives policy-makers an evidence base and context to ensure that patent practice is aligned with the economic and social goals of the IP system. In each of these critical roles, patent office practice in relation to gene patenting falls short.
DNA and protein sequences are made up of either combinations of four letters – A, C, G, and T, in the case of DNA – or 20 types of amino acids, each with different chemical properties – in the case of protein. To understand their structure, function and similarity, they must be read using specialized computer software tools. Therefore, when disclosed in a patent document, an examiner or practitioner would need to use computer-mediated searching, analysis and visual tools to interpret their contextual value or meaning. Any form used in the patenting process that does not facilitate access to computer searchable information – often called ‘machine readable’ - in our view fails to meet the patent system’s public disclosure requirements.
In a recent publication we reported the development and availability of an open, public, web-based platform that has great potential to improve the transparency of gene patenting worldwide.
Our international survey of 55 patent offices during the period July to October 2011 which focused on standards and practices regarding patentability of genetic sequences, revealed significant room for improvement in terms of making these data freely and publicly available for aggregation. While patentability requirements appear to be converging in at least 35 respondent patent offices, public disclosure of patent sequences remains restricted to visual inspection. Open public capabilities to search and analyse sequence-based discoveries and inventions are almost non-existent.
To address this gap, Cambia has expanded the publicly searchable patent sequence database in The Lens to include data from 15 jurisdictions and has developed a suite of new patent sequence (PatSeq) tools to enable exploration of the legal and scientific information within biological patents as they relate to a particular genome. The first exploratory tools target gene patents and their disclosed biological sequences associated with the human genome, and provide a platform to map, analyse, annotate and share this knowledge with anyone.
Although the recent decision by the US Supreme Court on breast cancer genes (Association for Molecular Pathology (AMP) v Myriad Genetics) held that naturally occurring sequences are not patentable in the US (see What Myriad means for biotech - WIPO Magazine), isolated genomic sequences are still patentable in many jurisdictions, including in Australia, Canada, Europe and Japan (for an example of a gene patent, check claims 1-3 in the Australian patent AU_686004_B2 at www.lens.org/lens/patent/AU_686004_B2).
As markets and innovation become increasingly globalized, such differences in national patent practice and policy underline the need for improved, standardized and open data sets, and improved compliance standards, as well as shared, open, decision-making tools to support the development of a favorable policy environment for biological innovation.
The Myriad case also highlighted the technical complexity of genomic sequence-based discoveries and inventions and the urgent need for more precise tools that identify similarities in the sequences disclosed in patents, especially in the claims section of a patent.
In gene patenting, there is a critical difference between disclosure of sequences and claiming of sequences. Upon submitting a patent application for a biological invention, the applicant is required to disclose all involved sequences (those that are simply used as references in the document, those that support the invention, or those that constitute the invention), in a separate section, called sequence listings section. When all these sequences are disclosed, it is critical to have tools that distinguish and illuminate the role, function, and location of each disclosed sequence vis-à-vis the invention, as well as its similarity to all previously disclosed sequences. Without such technical knowledge and clarifying tools, the public, many policy-makers, innovators and investors are often confused about the extent and scope of gene patents.
In general, we found that many sequences are disclosed but few are claimed as genes. If a sequence per se is claimed, it is usually claimed as an isolated or purified molecule in a particular jurisdiction. A claimed gene sequence means that any potential use of that sequence is restricted and will need to be licensed from the patent holder for the duration of the patent term or for as long as the patent is active in that jurisdiction. However, if the sequence is claimed as part of a larger sequence or as a target for a specific method, the uses of that particular sequence are unlikely to be exclusive making it possible for other inventors to access and use it freely without the need to negotiate a license. The public and innovators must be able to readily distinguish these cases to better understand the extent and scope of granted rights on gene sequences, reduce investment risks, and stimulate an equitable and inclusive innovation system, but this is extremely difficult.
While major patent offices claim to have sophisticated databases available to them that comprise a substantial set of sequences, in general the public cannot access or use these tools. Moreover, many patent offices with limited budgets or serving jurisdictions with emerging IP systems do not have access to such tools.
Even informal collaborations that seek to harmonize patent sequence disclosure and availability between countries are limited in scope. For example, the collaboration between the DNA Databank of Japan (DDBJ); the European Nucleotide Archives (ENA); and the GenBank-PAT division at the National Center for Biotechnology Information (NCBI) in the US, is limited to sharing nucleotide-based patent sequences with no formal agreement, as yet, to share protein-based patent sequences. The International Nucleotide Sequence Database Collaboration (INSDC), which brings these three major public databases together, fosters the exchange of nucleotide-based patent sequences on a daily basis. While each database may have duplicate sequence listings from PCT applications and granted US patents, they each maintain a slightly different record of patent sequences. While some commercial vendors claim to offer comprehensive data and sophisticated analysis, this is an expensive means of accessing public information. And even these commercial databases are incomplete.
Patent-disclosed sequences and the human genome
Cambia’s biological facility within The Lens provides an evidence-based understanding of the complex gene patenting landscape. It allows sequence and patent data aggregation, analysis and visualization, and is equipped with tools to dynamically search and find patent sequences associated with several genomes with various degrees of similarity, and to compare the scope of patenting, beginning with the human genome. Our analysis of the scope of patenting of known genes on the human genome showed that the percentage of known genes referenced – mentioned in the claims section of the patent but not necessarily claimed - ranges from 26 percent to 62 percent.
By August 2012, we had 131,339 nucleotide and 15,054 amino acid sequence listing entries mapped with 100 percent homology onto the human genome. These were referenced in the claims of 13,985 US issued patents.
Claimed versus disclosed nucleotide sequences
After optimizing and extending the algorithms to select patent documents that reference a sequence in their claims, we began analyzing manually the claims associated with only the fully aligned 131,339 nucleotide-sequence entries (not the amino acid sequence entries). These were referenced in 2,716 patents. We found that 76,910 sequences out of the 131,339 sequences mapped uniquely with 100 percent homology to the human genome and corresponded to 2,685 patents. The remaining 54,429 sequences were repeated in duplicated patent documents. According to the Myriad decision, the fully aligned sequences would be considered natural nucleotide sequences from the human genome, and therefore non-patentable in an unmodified form.
An analysis of the claims referencing these unique sequences revealed a variety of strategies for claiming a nucleotide sequence; that a small proportion (13 percent) of these sequences are claimed as sequence composition (having exclusive rights on the use of that sequence); and that about a third of the corresponding patents were not maintained for their full potential lifetime (i.e. 20 years). Further analysis of the claimed and patented nucleotide sequences suggested that the public and private institutes have different perceptions of the value of gene patents and the models for their use.
The PatSeq Toolkit
In the multilingual Lens facility, we developed a suite of tools to navigate patent and sequence information. For example, when a patent document contains a sequence disclosure, a small helical DNA structure appears in the information column of the search results page alerting users to its availability. We have also introduced a sequence tab that clarifies the nature of the disclosed sequence(s) within the document portfolio allowing users to search and filter the metadata (nature of sequence, length, origin of organism), whenever available, to locate the sequence within the document, and to view the original data source (where we downloaded the sequence from). We also created PatSeq Explorer, PatSeq Analyzer, and PatSeq Finder for more in-depth analyses.
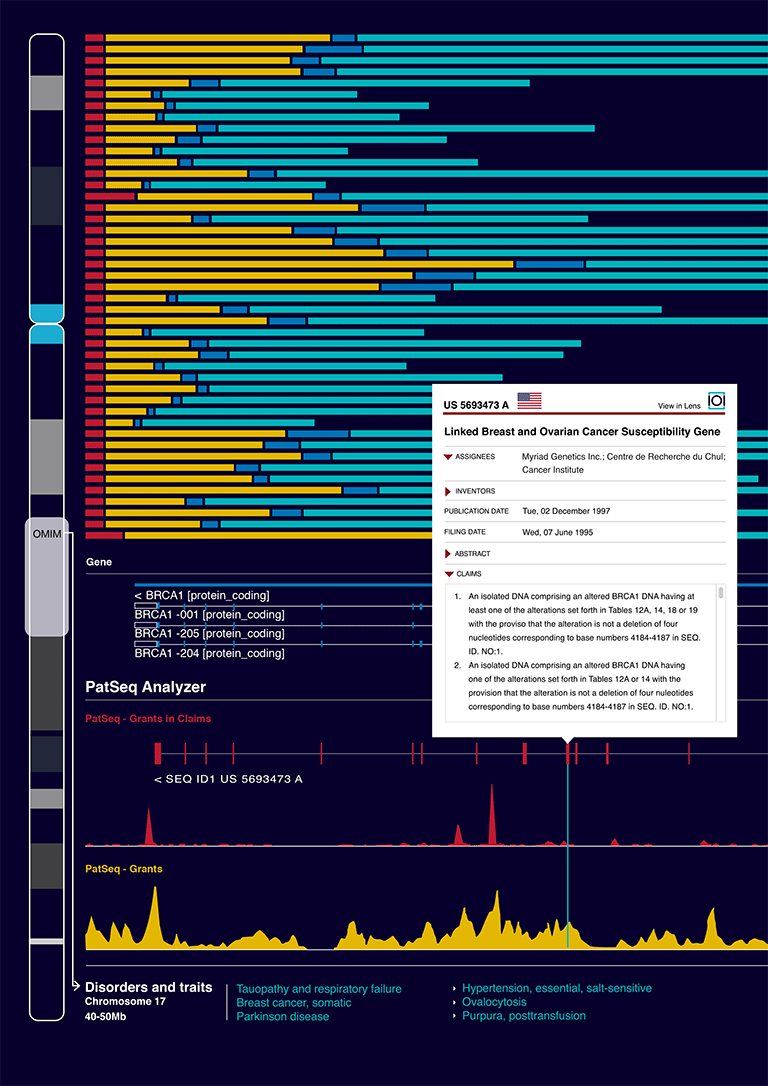
PatSeq Explorer enables multi-level visualization and navigation of patent disclosed sequences that map according to various homology thresholds to a reference genome, the first publicly available example of which is the human genome. At the genome and chromosome levels, users can investigate overall patenting trends, filter, and search sequence and patent attributes, and link to various sets of patent documents in The Lens database or choose to investigate further and analyze the sequence at the locus and gene levels (see Fig. 1). Mapped sequence entries are displayed according to their location in the patent document and their type, along with a summary panel view of their numbers and their corresponding patent documents counts in the jurisdictions in which the sequences were disclosed. All views are embeddable in blogs and social network facilities (to encourage uptake of evidence-based tools) and we expect all documents and sequence collections to be downloadable in the near future.
PatSeq Analyzer enables users to zoom in to the details of a particular sequence entry, view and compare disclosed sequence ID numbers within and across various patent documents and analyze their corresponding patent attributes within a specific gene area. The tool is a modified genome viewer built and integrated into PatSeq Explorer based on the open source HTML5/SVG genome maps browser developed by the Computational Medicine Institute, Prince Felipe Research Centre, Valencia, Spain. All views in PatSeq Analyzer are also embeddable.
PatSeq Finder enables users to query their own sequence against the PatSeq databases and conduct sequence similarity searches. Results from such searches are aligned based on a score of relatedness to the original query sequence and display information relating to corresponding patent documents. Users can view sequences referenced in the claims, read the corresponding patent claims, examine alignment details, sequence annotation, and embed or download results in various formats.
Patent office survey results
Almost all respondent patent offices indicated that they comply with the agreed standard for disclosure of sequences associated with patent filings (the ST.25 standard), which unfortunately, does not stipulate machine-readability. Most offices - with the exception of Israel - make sequence listings publicly available. However, they are available mostly as part of the published patent document, in pdf or image formats, and thus not machine-readable. A few patent offices, such as those of Canada, Germany, and Hungary provide machine-searchable sequence listings on their websites, whereas Japan, the EPO, and the US, as well as the Republic of Korea to some extent, provide machine-searchable sequence listings through third party providers or electronic downloadable files via their websites. These are often fee-based.
While the survey generated a more realistic picture of the total count of sequence listings from some jurisdictions, this proved difficult for many others, especially those that rely on regional patent offices, such as the EPO, and WIPO, for that information, or those that do not disclose publicly the yearly counts of such sequence listings. For example, in the US, where compliance with sequence rules is more rigorously observed, we found several thousand sequence listings cited in patents published since 1990 that were not included in the GenBank–PAT division database.
Our survey confirmed the lack of transparent public tools to navigate gene patents. While the Myriad decision has clarified, to some extent, the US position on gene sequence claiming, the ruling also exposed the critical need for, and lack of, nuanced and precise analysis of gene patents at both national and global level. Without transparency tools, the public will be disadvantaged, uncertainty will continue, compromising entrepreneurialism and investment, and inefficient use of resources will persist in industry and public sector innovation, to the detriment of informed policy making. Cambia’s biological facility within The Lens offers an open public platform that serves as a uniquely valuable alternative to the current commercial services that serve only a few elite innovators in biological sciences.
The author acknowledges the contributions of the co-authors in the Nature Biotech article.
About Cambia
Cambia, meaning change, is an independent non-profit institute creating new technologies, tools and paradigms to promote change and innovation. Its mission is to democratize innovation: to create a more equitable and inclusive capability to solve global problems using science and technology.